ORR Go/Nogo Design
ORR_Gonogo.Rmd
library(gonogo)
library(ggplot2)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, unionBasic Settings
In this article, an example of Go/Nogo design based on ORR is demonstrated. First we need some settings:
nmax <- 40
n1 <- 20
eff_cut <- 0.55
fut_cut <- 0.45
fa_eff_pt <- 0.6
fa_fut_pt <- 0.5- The total sample size at final analysis is 40.
- The sample size at interim analysis is 20.
- The efficacy boundary of ORR is 0.55.
- The futility boundary of ORR is 0.45.
- The (posterior) probability target/threshold of efficacy is 0.6. We declare efficacy if Pr(ORR > 0.55 | Data) > 0.6.
- The (posterior) probability target/threshold of futility is 0.5. We declare futility if Pr(ORR < 0.45 | Data) > 0.5.
About our hypothetical product, we have the following information:
tpp_tb <- tibble::tribble(
~theta, ~tpp,
0.45, "SOC",
0.55, "TPP base",
0.60, "TPP best"
)
theta_pick <- c(0.45, 0.5, 0.55, 0.6) # theta(underlying ORR) we might be interestedSo the efficacy of current SOC is 0.45, our TPP base is 0.55 and TPP best is 0.6.
Interim Analysis
We can list all possible results at interim analysis:
m_vec <- 0 : n1
res <- BM_Res(
nmax = nmax, n1 = n1, m = m_vec,
pt = fa_eff_pt, theta0 = eff_cut,
eff_cut = eff_cut, fut_cut = fut_cut,
a = 1, b = 1
)
knitr::kable(format(res, visable = FALSE))| nmax | n1 | ORR | CI | Post_mean | HDI | Go_pprob | Nogo_pprob | Pred_prob |
|---|---|---|---|---|---|---|---|---|
| 40 | 20 | 0(0.00) | 0.00 - 0.17 | 0.05 | 0.00 - 0.13 | 0.0% | 100.0% | 0.0% |
| 40 | 20 | 1(0.05) | 0.00 - 0.25 | 0.09 | 0.00 - 0.21 | 0.0% | 100.0% | 0.0% |
| 40 | 20 | 2(0.10) | 0.01 - 0.32 | 0.14 | 0.02 - 0.28 | 0.0% | 99.9% | 0.0% |
| 40 | 20 | 3(0.15) | 0.03 - 0.38 | 0.18 | 0.04 - 0.34 | 0.0% | 99.7% | 0.0% |
| 40 | 20 | 4(0.20) | 0.06 - 0.44 | 0.23 | 0.07 - 0.40 | 0.1% | 98.7% | 0.0% |
| 40 | 20 | 5(0.25) | 0.09 - 0.49 | 0.27 | 0.10 - 0.46 | 0.4% | 96.1% | 0.0% |
| 40 | 20 | 6(0.30) | 0.12 - 0.54 | 0.32 | 0.13 - 0.51 | 1.3% | 90.4% | 0.0% |
| 40 | 20 | 7(0.35) | 0.15 - 0.59 | 0.36 | 0.17 - 0.56 | 3.8% | 80.3% | 0.3% |
| 40 | 20 | 8(0.40) | 0.19 - 0.64 | 0.41 | 0.21 - 0.61 | 9.1% | 65.9% | 1.9% |
| 40 | 20 | 9(0.45) | 0.23 - 0.68 | 0.45 | 0.25 - 0.66 | 18.4% | 48.8% | 7.5% |
| 40 | 20 | 10(0.50) | 0.27 - 0.73 | 0.50 | 0.30 - 0.70 | 32.1% | 32.1% | 21.0% |
| 40 | 20 | 11(0.55) | 0.32 - 0.77 | 0.55 | 0.34 - 0.75 | 48.8% | 18.4% | 43.0% |
| 40 | 20 | 12(0.60) | 0.36 - 0.81 | 0.59 | 0.39 - 0.79 | 65.9% | 9.1% | 67.4% |
| 40 | 20 | 13(0.65) | 0.41 - 0.85 | 0.64 | 0.44 - 0.83 | 80.3% | 3.8% | 86.0% |
| 40 | 20 | 14(0.70) | 0.46 - 0.88 | 0.68 | 0.49 - 0.87 | 90.4% | 1.3% | 95.7% |
| 40 | 20 | 15(0.75) | 0.51 - 0.91 | 0.73 | 0.54 - 0.90 | 96.1% | 0.4% | 99.1% |
| 40 | 20 | 16(0.80) | 0.56 - 0.94 | 0.77 | 0.60 - 0.93 | 98.7% | 0.1% | 99.9% |
| 40 | 20 | 17(0.85) | 0.62 - 0.97 | 0.82 | 0.66 - 0.96 | 99.7% | 0.0% | 100.0% |
| 40 | 20 | 18(0.90) | 0.68 - 0.99 | 0.86 | 0.72 - 0.98 | 99.9% | 0.0% | 100.0% |
| 40 | 20 | 19(0.95) | 0.75 - 1.00 | 0.91 | 0.79 - 1.00 | 100.0% | 0.0% | 100.0% |
| 40 | 20 | 20(1.00) | 0.83 - 1.00 | 0.95 | 0.87 - 1.00 | 100.0% | 0.0% | 100.0% |
In this table, we have number of observed responders, observed ORR, 95% CI based on Clopper-Pearson method, posterior mean of ORR, 95% HDI of ORR, posterior probability of efficacy given current data, posterior probability of futility given current data, and the predictive probability of “Declare efficacy at FA” given current data.
ia_eff_pt <- 0.5
ia_fut_pt <- 0.01
ia_rules <- Find_Cut(
nmax = nmax, n1 = n1, m = m,
pt = fa_eff_pt, theta0 = eff_cut,
eff_cut = eff_cut, fut_cut = fut_cut,
eff_pt = ia_eff_pt, fut_pt = ia_fut_pt,
a = 1, b = 1
)If we set the rules to Go if predictive probability greater than 0.5 and to Nogo if predictive probability less than 0.01. Then we can determine the cut value for Go/Nogo as:
- Declare Go if #resp is equal to or greater than 12, which means observed ORR >= 0.6. The predictive probability of “Pr(ORR > 0.55 | Data) > 0.6 at 40 subjects” is 67.4%.
- Declare Nogo if #resp is equal to or less than 7, which means observed ORR <= 0.35. The predictive probability of “Pr(ORR > 0.55 | Data) > 0.6 at 40 subjects” is 0.3%.
Given this rule, we can compute the operating characteristics (OC) table for various observed ORR
oc_tab_s1 <- t(sapply(theta_pick, function(orr, n, go_cut, nogo_cut){
res <- ORR_Go_Nogo(n = n, orr = orr, go_cut = go_cut, nogo_cut = nogo_cut)
res <- format(res, visable = FALSE)
},
n = n1,
go_cut = as.integer(ia_rules["go_cut"]),
nogo_cut = as.integer(ia_rules["nogo_cut"])))
knitr::kable(oc_tab_s1)| Underlying_ORR | Pr_Go | Pr_Nogo | Pr_Equivocal |
|---|---|---|---|
| 45.00% | 13.1% | 25.2% | 61.7% |
| 50.00% | 25.2% | 13.2% | 61.7% |
| 55.00% | 41.4% | 5.8% | 52.8% |
| 60.00% | 59.6% | 2.1% | 38.3% |
We can also show OC figures of different rules
rules <- data.frame(
gocut = c(ia_rules["go_cut"] - 1, ia_rules["go_cut"] + 1),
nogocut = c(ia_rules["nogo_cut"] + 1, ia_rules["nogo_cut"] - 1),
n = n1
) %>%
mutate(nogocut = if_else(
nogocut < gocut, nogocut, gocut - 1
)) %>%
bind_rows(
data.frame(
gocut = ia_rules["go_cut"],
nogocut = ia_rules["nogo_cut"],
n = n1)) %>%
distinct() %>%
arrange(gocut)
oc_fig_s1 <- OC_Curve_Diff_Rules(rules, theta_pick,
tpp_tb = tpp_tb,
plot_params = list(
contour_vec = seq(from = 0.25, to = 1, by = 0.25)))
oc_fig_s1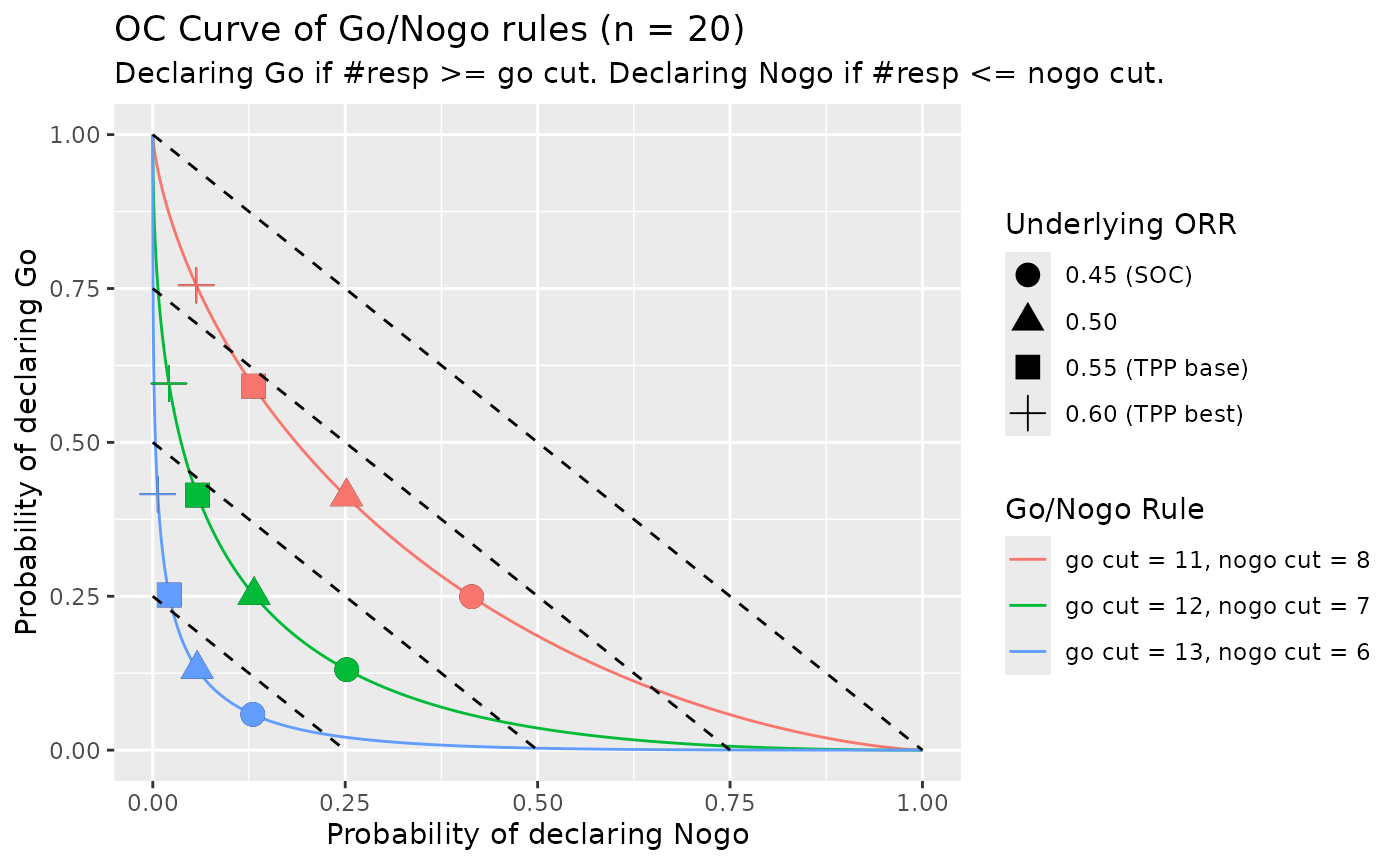
Full Monitoring Table
Based on Predictive Probability
enroll_num_vec <- 1 : n1
res <- matrix(NA, nrow = n1 + 1, ncol = n1)
colnames(res) <- paste0("enroll_", seq(n1))
rownames(res) <- paste0("#resp_", seq(n1 + 1) - 1)
for(enroll_num in seq(n1)){
for(row_idx in seq(from = 1, to = enroll_num + 1)){
resp_num <- row_idx - 1
pred_prob <- gonogo:::pred_prob2(nmax = nmax, n1 = enroll_num, m = resp_num,
pt = fa_eff_pt, theta0 = eff_cut)
res[row_idx, enroll_num] <- pred_prob
}
}
knitr::kable(round(res, digits = 3))| enroll_1 | enroll_2 | enroll_3 | enroll_4 | enroll_5 | enroll_6 | enroll_7 | enroll_8 | enroll_9 | enroll_10 | enroll_11 | enroll_12 | enroll_13 | enroll_14 | enroll_15 | enroll_16 | enroll_17 | enroll_18 | enroll_19 | enroll_20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #resp_0 | 0.187 | 0.077 | 0.030 | 0.011 | 0.004 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| #resp_1 | 0.691 | 0.407 | 0.216 | 0.105 | 0.048 | 0.020 | 0.008 | 0.003 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| #resp_2 | NA | 0.834 | 0.598 | 0.381 | 0.220 | 0.117 | 0.057 | 0.026 | 0.011 | 0.004 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| #resp_3 | NA | NA | 0.913 | 0.742 | 0.542 | 0.358 | 0.216 | 0.120 | 0.061 | 0.029 | 0.012 | 0.005 | 0.002 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| #resp_4 | NA | NA | NA | 0.955 | 0.843 | 0.679 | 0.500 | 0.336 | 0.208 | 0.118 | 0.061 | 0.029 | 0.013 | 0.005 | 0.002 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 |
| #resp_5 | NA | NA | NA | NA | 0.978 | 0.908 | 0.787 | 0.630 | 0.465 | 0.316 | 0.197 | 0.113 | 0.059 | 0.028 | 0.012 | 0.005 | 0.002 | 0.000 | 0.000 | 0.000 |
| #resp_6 | NA | NA | NA | NA | NA | 0.989 | 0.949 | 0.865 | 0.741 | 0.589 | 0.434 | 0.295 | 0.185 | 0.106 | 0.055 | 0.026 | 0.011 | 0.004 | 0.001 | 0.000 |
| #resp_7 | NA | NA | NA | NA | NA | NA | 0.995 | 0.972 | 0.919 | 0.827 | 0.700 | 0.553 | 0.406 | 0.275 | 0.171 | 0.097 | 0.049 | 0.023 | 0.009 | 0.003 |
| #resp_8 | NA | NA | NA | NA | NA | NA | NA | 0.998 | 0.986 | 0.953 | 0.890 | 0.792 | 0.664 | 0.521 | 0.379 | 0.254 | 0.156 | 0.086 | 0.043 | 0.019 |
| #resp_9 | NA | NA | NA | NA | NA | NA | NA | NA | 0.999 | 0.993 | 0.975 | 0.934 | 0.863 | 0.760 | 0.631 | 0.490 | 0.352 | 0.233 | 0.140 | 0.075 |
| #resp_10 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 0.997 | 0.987 | 0.962 | 0.914 | 0.837 | 0.730 | 0.600 | 0.460 | 0.326 | 0.210 |
| #resp_11 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 0.999 | 0.994 | 0.980 | 0.950 | 0.896 | 0.813 | 0.702 | 0.570 | 0.430 |
| #resp_12 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 0.999 | 0.997 | 0.990 | 0.972 | 0.937 | 0.878 | 0.790 | 0.674 |
| #resp_13 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 1.000 | 0.999 | 0.995 | 0.985 | 0.964 | 0.925 | 0.860 |
| #resp_14 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 1.000 | 0.999 | 0.998 | 0.993 | 0.981 | 0.957 |
| #resp_15 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 1.000 | 1.000 | 0.999 | 0.997 | 0.991 |
| #resp_16 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 1.000 | 1.000 | 1.000 | 0.999 |
| #resp_17 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 1.000 | 1.000 | 1.000 |
| #resp_18 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 1.000 | 1.000 |
| #resp_19 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 1.000 |
| #resp_20 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 |
Based on Posterior Probability
eff_res <- matrix(NA, nrow = n1 + 1, ncol = n1)
colnames(eff_res) <- paste0("enroll_", seq(n1))
rownames(eff_res) <- paste0("#resp_", seq(n1 + 1) - 1)
fut_res <- matrix(NA, nrow = n1 + 1, ncol = n1)
colnames(fut_res) <- paste0("enroll_", seq(n1))
rownames(fut_res) <- paste0("#resp_", seq(n1 + 1) - 1)
for(enroll_num in seq(n1)){
for(row_idx in seq(from = 1, to = enroll_num + 1)){
resp_num <- row_idx - 1
post_prob <- gonogo:::IA_post(n = enroll_num, m = resp_num,
eff_cut = eff_cut, fut_cut = fut_cut)
eff_res[row_idx, enroll_num] <- post_prob[1]
fut_res[row_idx, enroll_num] <- post_prob[2]
}
}Posterior probability for efficacy: Pr(ORR > 0.55| Data)
| enroll_1 | enroll_2 | enroll_3 | enroll_4 | enroll_5 | enroll_6 | enroll_7 | enroll_8 | enroll_9 | enroll_10 | enroll_11 | enroll_12 | enroll_13 | enroll_14 | enroll_15 | enroll_16 | enroll_17 | enroll_18 | enroll_19 | enroll_20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #resp_0 | 0.202 | 0.091 | 0.041 | 0.018 | 0.008 | 0.004 | 0.002 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| #resp_1 | 0.698 | 0.425 | 0.241 | 0.131 | 0.069 | 0.036 | 0.018 | 0.009 | 0.005 | 0.002 | 0.001 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| #resp_2 | NA | 0.834 | 0.609 | 0.407 | 0.255 | 0.153 | 0.088 | 0.050 | 0.027 | 0.015 | 0.008 | 0.004 | 0.002 | 0.001 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| #resp_3 | NA | NA | 0.908 | 0.744 | 0.558 | 0.392 | 0.260 | 0.166 | 0.102 | 0.061 | 0.036 | 0.020 | 0.011 | 0.006 | 0.003 | 0.002 | 0.001 | 0.001 | 0.000 | 0.000 |
| #resp_4 | NA | NA | NA | 0.950 | 0.836 | 0.684 | 0.523 | 0.379 | 0.262 | 0.174 | 0.112 | 0.070 | 0.043 | 0.025 | 0.015 | 0.009 | 0.005 | 0.003 | 0.002 | 0.001 |
| #resp_5 | NA | NA | NA | NA | 0.972 | 0.898 | 0.780 | 0.639 | 0.496 | 0.367 | 0.261 | 0.179 | 0.119 | 0.077 | 0.049 | 0.030 | 0.018 | 0.011 | 0.006 | 0.004 |
| #resp_6 | NA | NA | NA | NA | NA | 0.985 | 0.937 | 0.850 | 0.734 | 0.603 | 0.473 | 0.356 | 0.259 | 0.182 | 0.124 | 0.083 | 0.054 | 0.034 | 0.021 | 0.013 |
| #resp_7 | NA | NA | NA | NA | NA | NA | 0.992 | 0.961 | 0.900 | 0.809 | 0.696 | 0.573 | 0.454 | 0.346 | 0.256 | 0.183 | 0.128 | 0.087 | 0.058 | 0.038 |
| #resp_8 | NA | NA | NA | NA | NA | NA | NA | 0.995 | 0.977 | 0.935 | 0.866 | 0.772 | 0.663 | 0.548 | 0.437 | 0.337 | 0.253 | 0.184 | 0.131 | 0.091 |
| #resp_9 | NA | NA | NA | NA | NA | NA | NA | NA | 0.997 | 0.986 | 0.958 | 0.907 | 0.833 | 0.739 | 0.634 | 0.526 | 0.422 | 0.329 | 0.249 | 0.184 |
| #resp_10 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.999 | 0.992 | 0.973 | 0.937 | 0.880 | 0.802 | 0.710 | 0.609 | 0.506 | 0.409 | 0.321 |
| #resp_11 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.999 | 0.995 | 0.983 | 0.958 | 0.915 | 0.853 | 0.774 | 0.683 | 0.586 | 0.488 |
| #resp_12 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 0.997 | 0.989 | 0.972 | 0.940 | 0.892 | 0.827 | 0.748 | 0.659 |
| #resp_13 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 0.998 | 0.993 | 0.982 | 0.959 | 0.922 | 0.870 | 0.803 |
| #resp_14 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 0.999 | 0.996 | 0.988 | 0.972 | 0.945 | 0.904 |
| #resp_15 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 0.999 | 0.997 | 0.992 | 0.981 | 0.961 |
| #resp_16 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 1.000 | 0.998 | 0.995 | 0.987 |
| #resp_17 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 1.000 | 0.999 | 0.997 |
| #resp_18 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 1.000 | 0.999 |
| #resp_19 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 | 1.000 |
| #resp_20 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.000 |
Posterior probability for futility: Pr(ORR <= 0.45| Data)
| enroll_1 | enroll_2 | enroll_3 | enroll_4 | enroll_5 | enroll_6 | enroll_7 | enroll_8 | enroll_9 | enroll_10 | enroll_11 | enroll_12 | enroll_13 | enroll_14 | enroll_15 | enroll_16 | enroll_17 | enroll_18 | enroll_19 | enroll_20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #resp_0 | 0.698 | 0.834 | 0.908 | 0.950 | 0.972 | 0.985 | 0.992 | 0.995 | 0.997 | 0.999 | 0.999 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| #resp_1 | 0.202 | 0.425 | 0.609 | 0.744 | 0.836 | 0.898 | 0.937 | 0.961 | 0.977 | 0.986 | 0.992 | 0.995 | 0.997 | 0.998 | 0.999 | 0.999 | 1.000 | 1.000 | 1.000 | 1.000 |
| #resp_2 | NA | 0.091 | 0.241 | 0.407 | 0.558 | 0.684 | 0.780 | 0.850 | 0.900 | 0.935 | 0.958 | 0.973 | 0.983 | 0.989 | 0.993 | 0.996 | 0.997 | 0.998 | 0.999 | 0.999 |
| #resp_3 | NA | NA | 0.041 | 0.131 | 0.255 | 0.392 | 0.523 | 0.639 | 0.734 | 0.809 | 0.866 | 0.907 | 0.937 | 0.958 | 0.972 | 0.982 | 0.988 | 0.992 | 0.995 | 0.997 |
| #resp_4 | NA | NA | NA | 0.018 | 0.069 | 0.153 | 0.260 | 0.379 | 0.496 | 0.603 | 0.696 | 0.772 | 0.833 | 0.880 | 0.915 | 0.940 | 0.959 | 0.972 | 0.981 | 0.987 |
| #resp_5 | NA | NA | NA | NA | 0.008 | 0.036 | 0.088 | 0.166 | 0.262 | 0.367 | 0.473 | 0.573 | 0.663 | 0.739 | 0.802 | 0.853 | 0.892 | 0.922 | 0.945 | 0.961 |
| #resp_6 | NA | NA | NA | NA | NA | 0.004 | 0.018 | 0.050 | 0.102 | 0.174 | 0.261 | 0.356 | 0.454 | 0.548 | 0.634 | 0.710 | 0.774 | 0.827 | 0.870 | 0.904 |
| #resp_7 | NA | NA | NA | NA | NA | NA | 0.002 | 0.009 | 0.027 | 0.061 | 0.112 | 0.179 | 0.259 | 0.346 | 0.437 | 0.526 | 0.609 | 0.683 | 0.748 | 0.803 |
| #resp_8 | NA | NA | NA | NA | NA | NA | NA | 0.001 | 0.005 | 0.015 | 0.036 | 0.070 | 0.119 | 0.182 | 0.256 | 0.337 | 0.422 | 0.506 | 0.586 | 0.659 |
| #resp_9 | NA | NA | NA | NA | NA | NA | NA | NA | 0.000 | 0.002 | 0.008 | 0.020 | 0.043 | 0.077 | 0.124 | 0.183 | 0.253 | 0.329 | 0.409 | 0.488 |
| #resp_10 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.000 | 0.001 | 0.004 | 0.011 | 0.025 | 0.049 | 0.083 | 0.128 | 0.184 | 0.249 | 0.321 |
| #resp_11 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.000 | 0.001 | 0.002 | 0.006 | 0.015 | 0.030 | 0.054 | 0.087 | 0.131 | 0.184 |
| #resp_12 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.000 | 0.000 | 0.001 | 0.003 | 0.009 | 0.018 | 0.034 | 0.058 | 0.091 |
| #resp_13 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.000 | 0.000 | 0.001 | 0.002 | 0.005 | 0.011 | 0.021 | 0.038 |
| #resp_14 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.000 | 0.000 | 0.000 | 0.001 | 0.003 | 0.006 | 0.013 |
| #resp_15 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.000 | 0.000 | 0.000 | 0.001 | 0.002 | 0.004 |
| #resp_16 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 |
| #resp_17 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.000 | 0.000 | 0.000 | 0.000 |
| #resp_18 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.000 | 0.000 | 0.000 |
| #resp_19 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.000 | 0.000 |
| #resp_20 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.000 |
Final Analysis
We can also list all possible results at final analysis
m_vec <- 0 : nmax
res <- BM_Res(nmax = nmax, n1 = nmax, m = m_vec,
pt = fa_eff_pt, theta0 = eff_cut,
eff_cut = eff_cut, fut_cut = fut_cut,
a = 1, b = 1)
knitr::kable(format(res, visable = FALSE))| nmax | n1 | ORR | CI | Post_mean | HDI | Go_pprob | Nogo_pprob | Pred_prob |
|---|---|---|---|---|---|---|---|---|
| 40 | 40 | 0(0.00) | 0.00 - 0.09 | 0.02 | 0.00 - 0.07 | 0.0% | 100.0% | NA |
| 40 | 40 | 1(0.03) | 0.00 - 0.13 | 0.05 | 0.00 - 0.11 | 0.0% | 100.0% | NA |
| 40 | 40 | 2(0.05) | 0.01 - 0.17 | 0.07 | 0.01 - 0.15 | 0.0% | 100.0% | NA |
| 40 | 40 | 3(0.07) | 0.02 - 0.20 | 0.10 | 0.02 - 0.18 | 0.0% | 100.0% | NA |
| 40 | 40 | 4(0.10) | 0.03 - 0.24 | 0.12 | 0.03 - 0.22 | 0.0% | 100.0% | NA |
| 40 | 40 | 5(0.12) | 0.04 - 0.27 | 0.14 | 0.05 - 0.25 | 0.0% | 100.0% | NA |
| 40 | 40 | 6(0.15) | 0.06 - 0.30 | 0.17 | 0.06 - 0.28 | 0.0% | 100.0% | NA |
| 40 | 40 | 7(0.17) | 0.07 - 0.33 | 0.19 | 0.08 - 0.31 | 0.0% | 100.0% | NA |
| 40 | 40 | 8(0.20) | 0.09 - 0.36 | 0.21 | 0.10 - 0.34 | 0.0% | 99.9% | NA |
| 40 | 40 | 9(0.23) | 0.11 - 0.38 | 0.24 | 0.12 - 0.37 | 0.0% | 99.8% | NA |
| 40 | 40 | 10(0.25) | 0.13 - 0.41 | 0.26 | 0.13 - 0.40 | 0.0% | 99.5% | NA |
| 40 | 40 | 11(0.28) | 0.15 - 0.44 | 0.29 | 0.15 - 0.42 | 0.0% | 98.7% | NA |
| 40 | 40 | 12(0.30) | 0.17 - 0.47 | 0.31 | 0.17 - 0.45 | 0.1% | 97.1% | NA |
| 40 | 40 | 13(0.33) | 0.19 - 0.49 | 0.33 | 0.19 - 0.48 | 0.2% | 94.1% | NA |
| 40 | 40 | 14(0.35) | 0.21 - 0.52 | 0.36 | 0.22 - 0.50 | 0.6% | 89.3% | NA |
| 40 | 40 | 15(0.38) | 0.23 - 0.54 | 0.38 | 0.24 - 0.53 | 1.4% | 82.3% | NA |
| 40 | 40 | 16(0.40) | 0.25 - 0.57 | 0.40 | 0.26 - 0.55 | 2.9% | 72.8% | NA |
| 40 | 40 | 17(0.42) | 0.27 - 0.59 | 0.43 | 0.28 - 0.58 | 5.7% | 61.5% | NA |
| 40 | 40 | 18(0.45) | 0.29 - 0.62 | 0.45 | 0.30 - 0.60 | 10.2% | 49.2% | NA |
| 40 | 40 | 19(0.47) | 0.32 - 0.64 | 0.48 | 0.33 - 0.63 | 16.9% | 36.9% | NA |
| 40 | 40 | 20(0.50) | 0.34 - 0.66 | 0.50 | 0.35 - 0.65 | 25.9% | 25.9% | NA |
| 40 | 40 | 21(0.53) | 0.36 - 0.68 | 0.52 | 0.37 - 0.67 | 36.9% | 16.9% | NA |
| 40 | 40 | 22(0.55) | 0.38 - 0.71 | 0.55 | 0.40 - 0.70 | 49.2% | 10.2% | NA |
| 40 | 40 | 23(0.57) | 0.41 - 0.73 | 0.57 | 0.42 - 0.72 | 61.5% | 5.7% | NA |
| 40 | 40 | 24(0.60) | 0.43 - 0.75 | 0.60 | 0.45 - 0.74 | 72.8% | 2.9% | NA |
| 40 | 40 | 25(0.62) | 0.46 - 0.77 | 0.62 | 0.47 - 0.76 | 82.3% | 1.4% | NA |
| 40 | 40 | 26(0.65) | 0.48 - 0.79 | 0.64 | 0.50 - 0.78 | 89.3% | 0.6% | NA |
| 40 | 40 | 27(0.68) | 0.51 - 0.81 | 0.67 | 0.52 - 0.81 | 94.1% | 0.2% | NA |
| 40 | 40 | 28(0.70) | 0.53 - 0.83 | 0.69 | 0.55 - 0.83 | 97.1% | 0.1% | NA |
| 40 | 40 | 29(0.72) | 0.56 - 0.85 | 0.71 | 0.58 - 0.85 | 98.7% | 0.0% | NA |
| 40 | 40 | 30(0.75) | 0.59 - 0.87 | 0.74 | 0.60 - 0.87 | 99.5% | 0.0% | NA |
| 40 | 40 | 31(0.78) | 0.62 - 0.89 | 0.76 | 0.63 - 0.88 | 99.8% | 0.0% | NA |
| 40 | 40 | 32(0.80) | 0.64 - 0.91 | 0.79 | 0.66 - 0.90 | 99.9% | 0.0% | NA |
| 40 | 40 | 33(0.82) | 0.67 - 0.93 | 0.81 | 0.69 - 0.92 | 100.0% | 0.0% | NA |
| 40 | 40 | 34(0.85) | 0.70 - 0.94 | 0.83 | 0.72 - 0.94 | 100.0% | 0.0% | NA |
| 40 | 40 | 35(0.88) | 0.73 - 0.96 | 0.86 | 0.75 - 0.95 | 100.0% | 0.0% | NA |
| 40 | 40 | 36(0.90) | 0.76 - 0.97 | 0.88 | 0.78 - 0.97 | 100.0% | 0.0% | NA |
| 40 | 40 | 37(0.93) | 0.80 - 0.98 | 0.90 | 0.82 - 0.98 | 100.0% | 0.0% | NA |
| 40 | 40 | 38(0.95) | 0.83 - 0.99 | 0.93 | 0.85 - 0.99 | 100.0% | 0.0% | NA |
| 40 | 40 | 39(0.97) | 0.87 - 1.00 | 0.95 | 0.89 - 1.00 | 100.0% | 0.0% | NA |
| 40 | 40 | 40(1.00) | 0.91 - 1.00 | 0.98 | 0.93 - 1.00 | 100.0% | 0.0% | NA |
Note: Here we list all possible results, even those should have been stopped at interim analysis.
fa_rules <- Find_Cut(
nmax = nmax, n1 = nmax, m = m,
pt = fa_eff_pt, theta0 = eff_cut,
eff_cut = eff_cut, fut_cut = fut_cut,
eff_pt = fa_eff_pt, fut_pt = fa_fut_pt,
a = 1, b = 1
)If we set the rules to Go if posterior Pr(ORR > 0.55) is greater than 0.6 and to Nogo if posterior Pr(ORR <= 0.45) is less than 0.5. Then we can determine the cut value for Go/Nogo as:
- Declare Go if #resp is equal to or greater than 23, which means observed ORR >= 0.58, posterior probability Pr(ORR > 0.55 | Data) >= 61.5%.
- Declare Nogo if #resp is equal to or less than 17 which means observed ORR <= 0.42, posterior probability Pr(ORR < 0.45 | Data) >= 61.5%.
Given this rule, we can compute the operating characteristics (OC) table for various observed ORR
oc_tab_s2 <- t(sapply(theta_pick, function(orr, n, go_cut, nogo_cut){
res <- ORR_Go_Nogo(n = n, orr = orr, go_cut = go_cut, nogo_cut = nogo_cut)
res <- format(res, visable = FALSE)
},
n = nmax,
go_cut = as.integer(fa_rules["go_cut"]),
nogo_cut = as.integer(fa_rules["nogo_cut"])))
knitr::kable(oc_tab_s2)| Underlying_ORR | Pr_Go | Pr_Nogo | Pr_Equivocal |
|---|---|---|---|
| 45.00% | 7.7% | 43.9% | 48.4% |
| 50.00% | 21.5% | 21.5% | 57.0% |
| 55.00% | 43.9% | 7.7% | 48.4% |
| 60.00% | 68.9% | 1.9% | 29.3% |
We can also show OC figures of different rules
rules <- data.frame(
gocut = c(fa_rules["go_cut"] - 1, fa_rules["go_cut"] + 1),
nogocut = c(fa_rules["nogo_cut"] + 1, fa_rules["nogo_cut"] - 1),
n = nmax
) %>%
mutate(nogocut = if_else(
nogocut < gocut, nogocut, gocut - 1
)) %>%
bind_rows(
data.frame(
gocut = fa_rules["go_cut"],
nogocut = fa_rules["nogo_cut"],
n = nmax)) %>%
distinct() %>%
arrange(gocut)
oc_fig_s2 <- OC_Curve_Diff_Rules(rules, theta_pick,
tpp_tb = tpp_tb,
plot_params = list(
contour_vec = seq(from = 0.25, to = 1, by = 0.25)))
oc_fig_s2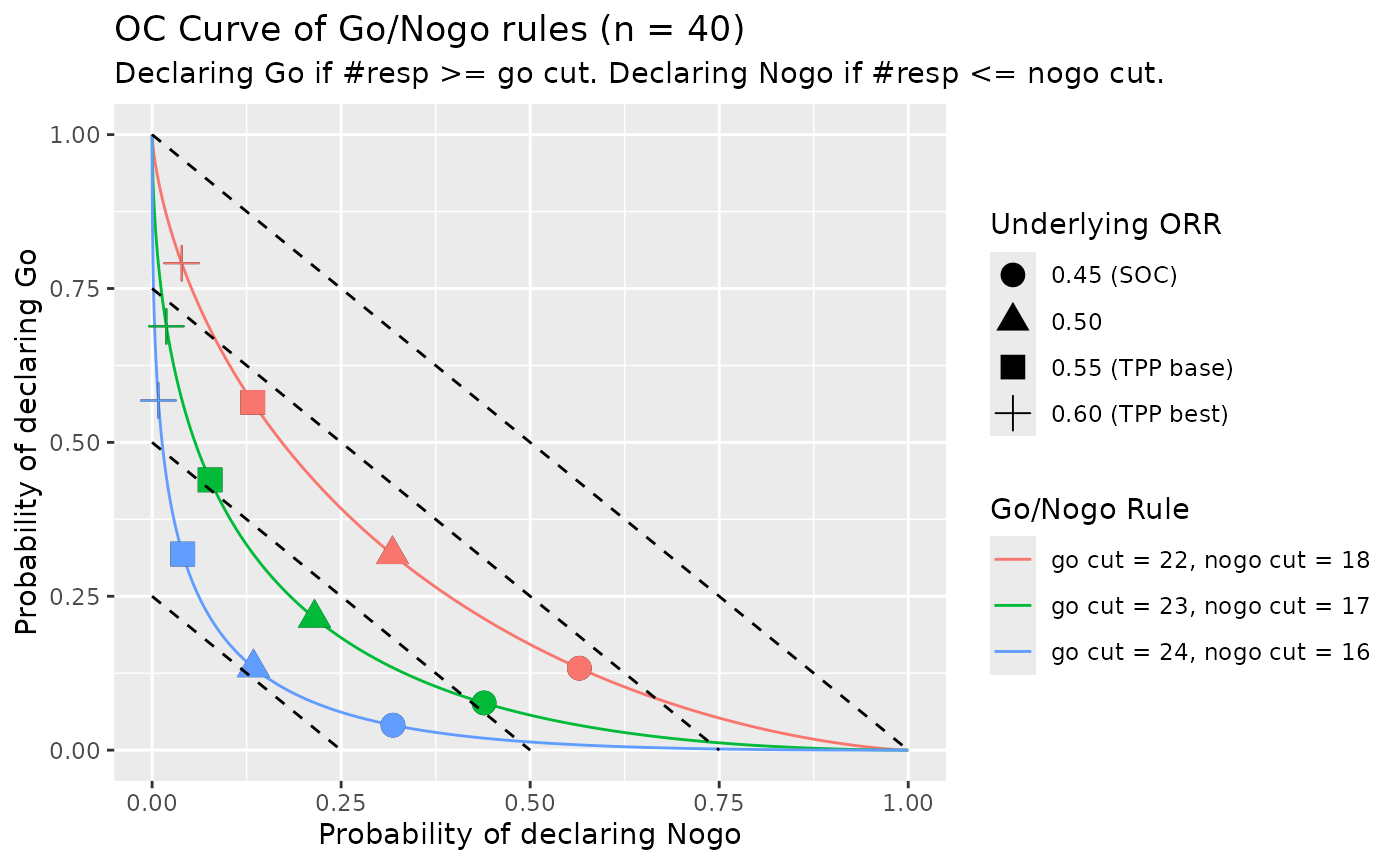
Figures for Confidence Given Different Rules
x <- seq(0, 1, by = 0.01)
# Nogo rules
res <- list()
for(i in fa_rules["nogo_cut"] + (-2 : 2)){
cdf <- pbeta(x, 1 + i, 1 + nmax - i, lower.tail = TRUE)
res <- append(res,
list(data.frame(ORR = x,
cdf = cdf,
resp = i)))
}
res <- do.call(rbind, res)
ggplot(res, aes(x = ORR, y = cdf, color = factor(resp))) +
geom_line() +
geom_vline(xintercept = fut_cut, linetype = "dashed") +
geom_hline(yintercept = fa_fut_pt, linetype = "dashed") +
labs(
x = "ORR futility cutoff",
y = "Pr(ORR <= Fut cut)",
color = "#resp") +
coord_cartesian(xlim = c(max(0, fut_cut - 0.3),
min(1, fut_cut + 0.3)))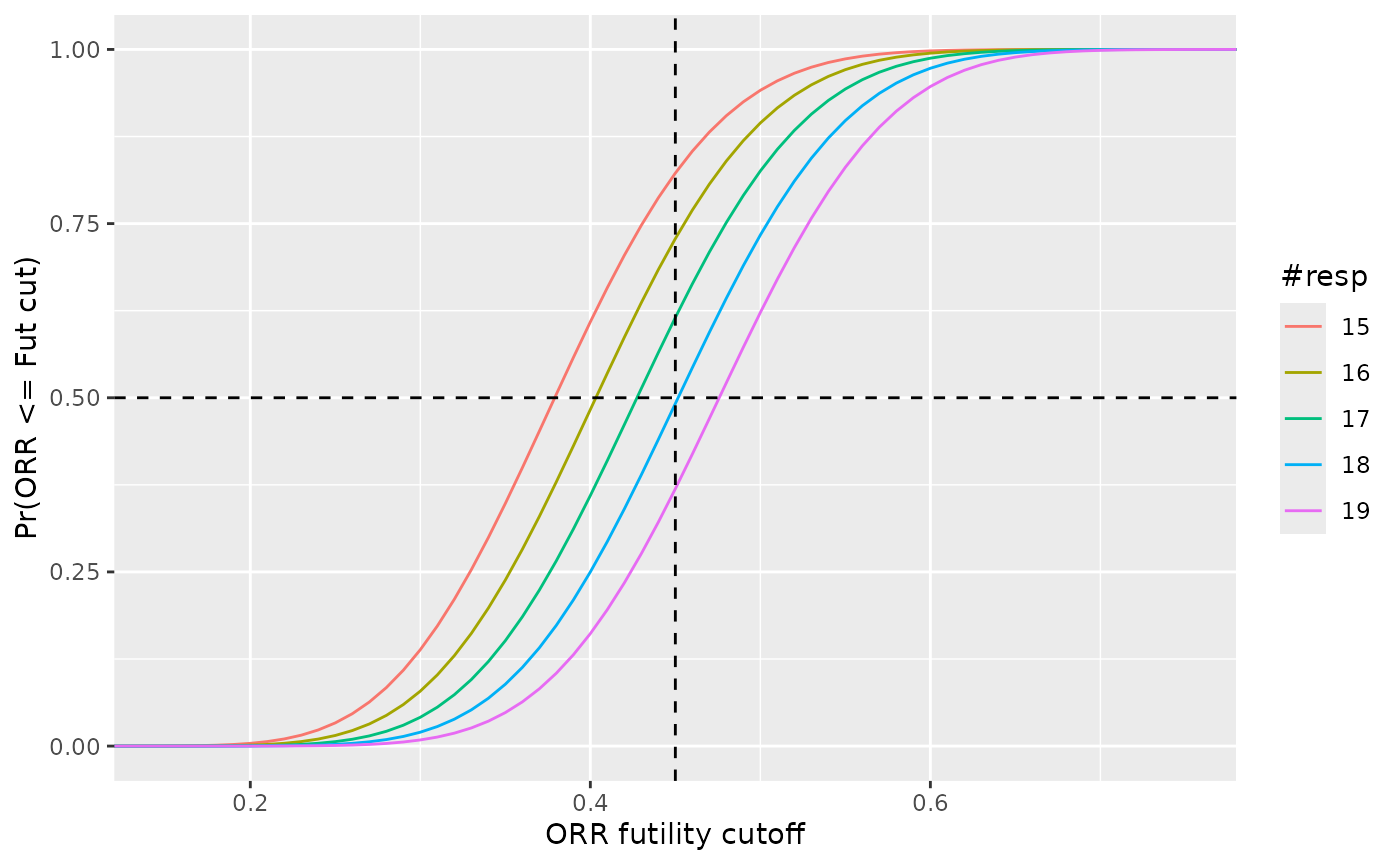
# Go rules
res <- list()
for(i in fa_rules["go_cut"] + (-2 : 2)){
cdf <- pbeta(x, 1 + i, 1 + nmax - i, lower.tail = FALSE)
res <- append(res,
list(data.frame(ORR = x,
cdf = cdf,
resp = i)))
}
res <- do.call(rbind, res)
ggplot(res, aes(x = ORR, y = cdf, color = factor(resp))) +
geom_line() +
geom_vline(xintercept = eff_cut, linetype = "dashed") +
geom_hline(yintercept = fa_eff_pt, linetype = "dashed") +
labs(
x = "ORR efficacy cutoff",
y = "Pr(ORR > Eff cut)",
color = "#resp") +
coord_cartesian(xlim = c(max(0, eff_cut - 0.3), min(1, eff_cut + 0.3)))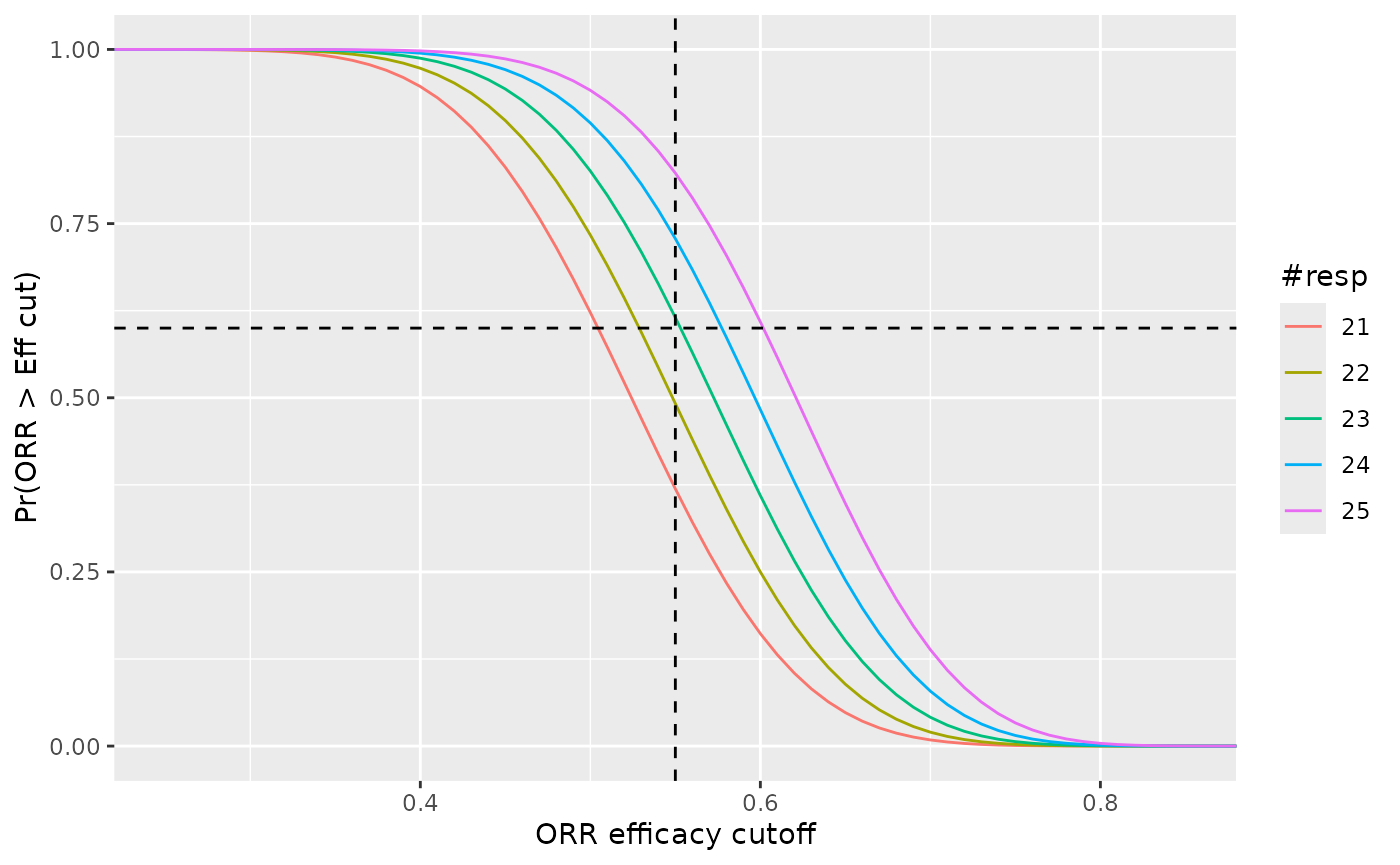
Appendix
Session info
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.4.1 (2024-06-14)
#> os Ubuntu 22.04.4 LTS
#> system x86_64, linux-gnu
#> ui X11
#> language en
#> collate C.UTF-8
#> ctype C.UTF-8
#> tz UTC
#> date 2024-06-24
#> pandoc 3.1.11 @ /opt/hostedtoolcache/pandoc/3.1.11/x64/ (via rmarkdown)
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> bayestestR 0.13.2 2024-02-12 [1] RSPM
#> bslib 0.7.0 2024-03-29 [1] RSPM
#> cachem 1.1.0 2024-05-16 [1] RSPM
#> cli 3.6.3 2024-06-21 [1] RSPM
#> colorspace 2.1-0 2023-01-23 [1] RSPM
#> desc 1.4.3 2023-12-10 [1] RSPM
#> digest 0.6.35 2024-03-11 [1] RSPM
#> dplyr * 1.1.4 2023-11-17 [1] RSPM
#> evaluate 0.24.0 2024-06-10 [1] RSPM
#> fansi 1.0.6 2023-12-08 [1] RSPM
#> farver 2.1.2 2024-05-13 [1] RSPM
#> fastmap 1.2.0 2024-05-15 [1] RSPM
#> fs 1.6.4 2024-04-25 [1] RSPM
#> generics 0.1.3 2022-07-05 [1] RSPM
#> ggplot2 * 3.5.1 2024-04-23 [1] RSPM
#> glue 1.7.0 2024-01-09 [1] RSPM
#> gonogo * 0.0.0.9024 2024-06-24 [1] local
#> gtable 0.3.5 2024-04-22 [1] RSPM
#> highr 0.11 2024-05-26 [1] RSPM
#> htmltools 0.5.8.1 2024-04-04 [1] RSPM
#> insight 0.20.1 2024-06-11 [1] RSPM
#> jquerylib 0.1.4 2021-04-26 [1] RSPM
#> jsonlite 1.8.8 2023-12-04 [1] RSPM
#> knitr 1.47 2024-05-29 [1] RSPM
#> labeling 0.4.3 2023-08-29 [1] RSPM
#> lifecycle 1.0.4 2023-11-07 [1] RSPM
#> magrittr 2.0.3 2022-03-30 [1] RSPM
#> memoise 2.0.1 2021-11-26 [1] RSPM
#> munsell 0.5.1 2024-04-01 [1] RSPM
#> pillar 1.9.0 2023-03-22 [1] RSPM
#> pkgconfig 2.0.3 2019-09-22 [1] RSPM
#> pkgdown 2.0.9 2024-04-18 [1] any (@2.0.9)
#> purrr 1.0.2 2023-08-10 [1] RSPM
#> quadprog 1.5-8 2019-11-20 [1] RSPM
#> R6 2.5.1 2021-08-19 [1] RSPM
#> ragg 1.3.2 2024-05-15 [1] RSPM
#> rlang 1.1.4 2024-06-04 [1] RSPM
#> rmarkdown 2.27 2024-05-17 [1] RSPM
#> sass 0.4.9 2024-03-15 [1] RSPM
#> scales 1.3.0 2023-11-28 [1] RSPM
#> sessioninfo 1.2.2 2021-12-06 [1] RSPM
#> stringi 1.8.4 2024-05-06 [1] RSPM
#> stringr 1.5.1 2023-11-14 [1] RSPM
#> systemfonts 1.1.0 2024-05-15 [1] RSPM
#> textshaping 0.4.0 2024-05-24 [1] RSPM
#> tibble 3.2.1 2023-03-20 [1] RSPM
#> tidyr 1.3.1 2024-01-24 [1] RSPM
#> tidyselect 1.2.1 2024-03-11 [1] RSPM
#> utf8 1.2.4 2023-10-22 [1] RSPM
#> vctrs 0.6.5 2023-12-01 [1] RSPM
#> withr 3.0.0 2024-01-16 [1] RSPM
#> xfun 0.45 2024-06-16 [1] RSPM
#> yaml 2.3.8 2023-12-11 [1] RSPM
#>
#> [1] /home/runner/work/_temp/Library
#> [2] /opt/R/4.4.1/lib/R/site-library
#> [3] /opt/R/4.4.1/lib/R/library
#>
#> ──────────────────────────────────────────────────────────────────────────────